The scikit-learn AUPRC implementation is wrong (or at least overly optimistic), apparently
Chen et al, fresh from the bioRxiv, write:
The precision-recall curve (PRC) and the area under it (AUPRC) are useful for quantifying classification performance. They are commonly used in situations with imbalanced classes, such as cancer diagnosis and cell type annotation. We evaluated 10 popular tools for plotting PRC and computing AUPRC, which were collectively used in >3,000 published studies. We found the AUPRC values computed by the tools rank classifiers differently and some tools produce overly-optimistic results.
There are, apparently, a handful of different ways to compute the “curves” for PRC and AUROC, which makes sense (given that they’re all built from discrete values), and furthermore, there is (per this paper) some consensus that certain methods are more “optimistic” than they should be. The authors go on to detail inconsistencies in method usage (and visualization) between core statistical libraries, including scikit-learn, with pretty striking results:
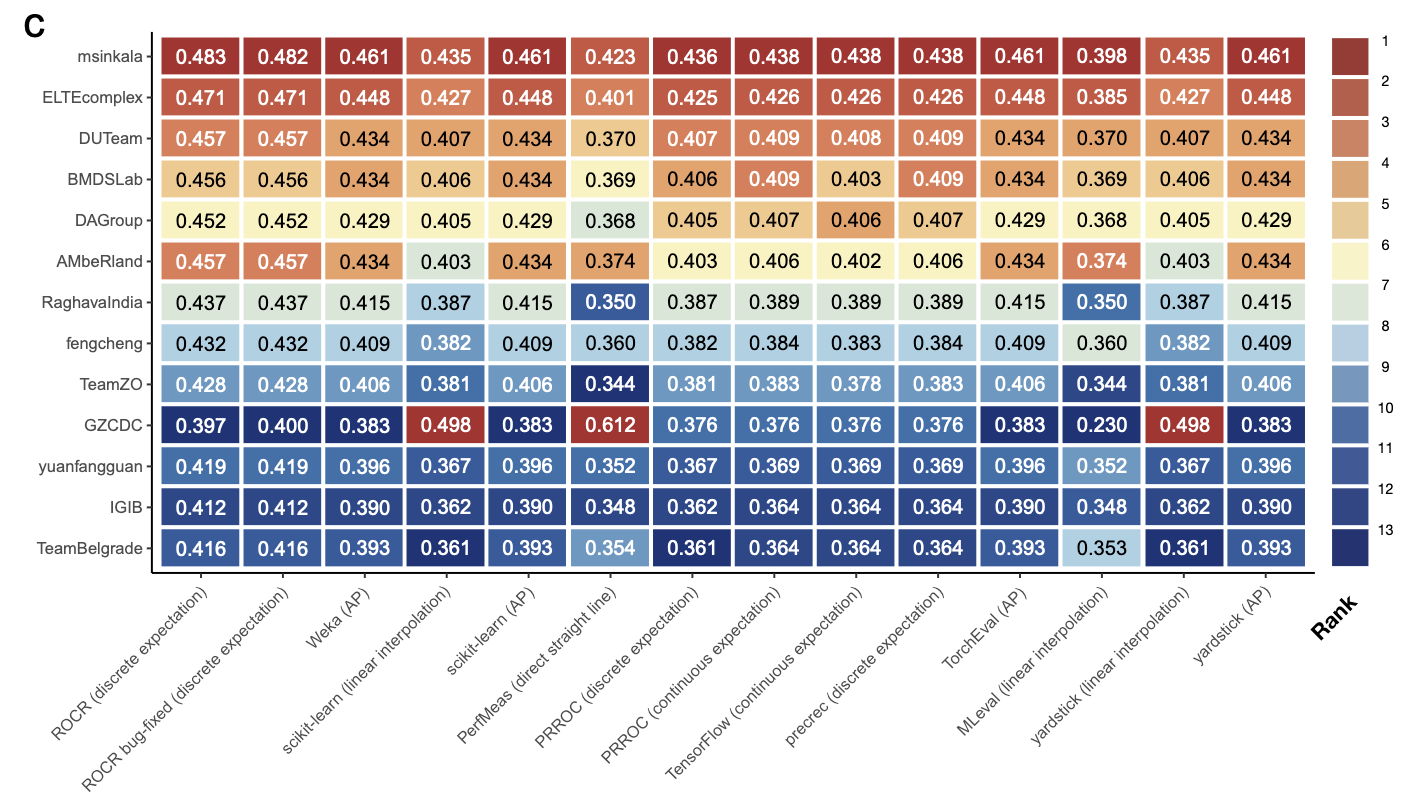
In some cases, these are pretty startling differences. They do note that, in the case of scikit-learn, the documentation at least notes the particulars of the method used, but personally, it wasn’t something I’d ever thought to pay attention to before.