Exploring the GSC 2024 Phenotyping Benchmark
A recent paper on a novel computational phenotyping method from Peter Robinson’s group includes a 2024 update to their GSC+ dataset, adding annotations from recent HPO updates to the original set of PubMed abstracts.
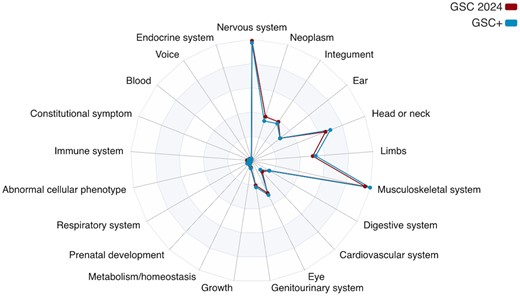
They helpfully include a figure describing the anatomical/system coverage of the provided annotations. The respective areas of coverage jump out – large swathes of the tree appear to be totally uncovered. This isn’t necessarily a bad thing, as the intention of the GSC corpus was to “match the 44 complex dysmorphology syndromes discussed in the initial HPO study”, per the original GSC paper. It does, of course, raise theoretical concerns about generalization, as other common phenotyping contexts might have a much broader mix of terms. Anecdotally, a first pass of terms employed in NICU notes here at Nationwide Children’s had much, much more even coverage overall, particularly of systems like blood, respiratory, and endocrine, and fewer neoplasm terms.
Using the eutils package, I downloaded metadata for each abstract and dug in a bit. Unsurprising given the content emphasis, many of the abstracts come from a small set of genetics journals:
| Journal name | # of abstracts |
|---|---|
| Am J Med Genet | 57 |
| J Med Genet | 23 |
| Am J Hum Genet | 21 |
| Am J Med Genet A | 11 |
| Clin Genet | 10 |
| Neurology | 7 |
| Hum Mol Genet | 7 |
| Hum Genet | 7 |
| Eur J Hum Genet | 6 |
| Genomics | 5 |
| Nature | 4 |
| Nat Genet | 4 |
| Lancet | 3 |
| Arch Dis Child | 3 |
| J Hum Genet | 3 |
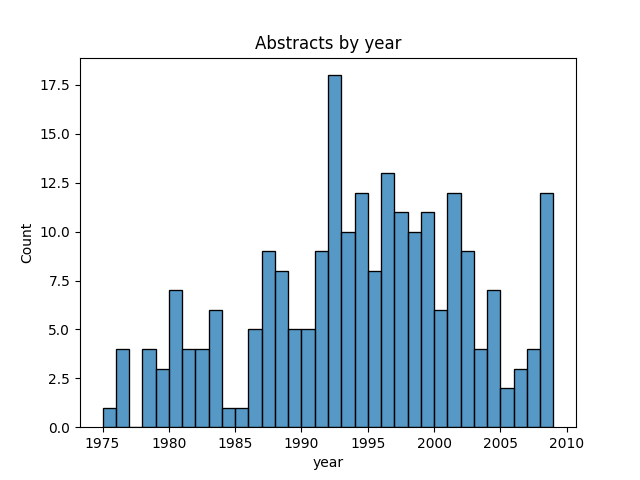
The abstracts go surprisingly far back. In discussing this, one of our scientists noted that a 1976 abstract uses the term “ankylois” that appears to be erroneous or anachronistic, and is a common failure point for most phenotyping tools. Because the abstract set was assembled several years ago, there’s nothing newer than 2008.
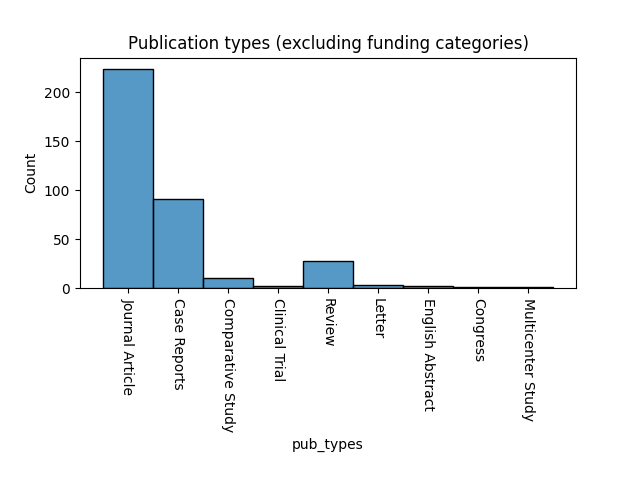
Included in the abstracts are a lot of case reports and reviews. Makes sense, but definitely a little different, contextually, than other cases like clinical notes.
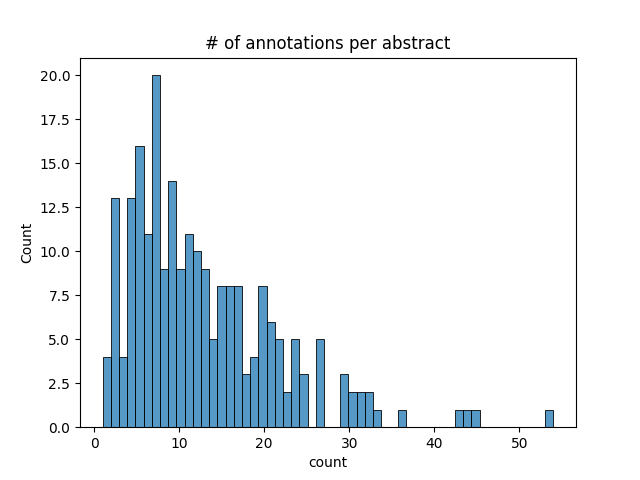
The density of annotations per abstracts varies considerably. Some are in the single digits. The median is probably somewhere around 10. A handful are above 40. Notably, from a QC perspective, there are none with zero (not the end of the world – there is plenty of text within the abstracts that works as a negative case).
Anyway, this was an enlightening exercise. I think these represent some potential minor limitations about the ability to generalize performance from how a phenotyping tool does against the GSC, but it’s hard to imagine that anyone will take the time to put something out in the form of e.g. anonymized annotated clinical notes.